Nicolaescu, L.: the Index of Families of Boundary Value Problems for Dirac Operators
Working with missing data¶
In this department, nosotros volition discuss missing (as well referred to as NA) values in pandas.
Notation
The selection of using NaN internally to denote missing data was largely for simplicity and performance reasons. Starting from pandas 1.0, some optional data types start experimenting with a native NA scalar using a mask-based arroyo. See hither for more.
Run into the cookbook for some avant-garde strategies.
Values considered "missing"¶
As data comes in many shapes and forms, pandas aims to be flexible with regard to handling missing information. While NaN is the default missing value marker for reasons of computational speed and convenience, we need to be able to easily detect this value with data of different types: floating betoken, integer, boolean, and full general object. In many cases, however, the Python None will arise and we wish to likewise consider that "missing" or "not available" or "NA".
Notation
If you want to consider inf and -inf to be "NA" in computations, you lot tin gear up pandas.options.mode.use_inf_as_na = True .
In [1]: df = pd . DataFrame ( ...: np . random . randn ( 5 , three ), ...: alphabetize = [ "a" , "c" , "e" , "f" , "h" ], ...: columns = [ "one" , "two" , "3" ], ...: ) ...: In [ii]: df [ "iv" ] = "bar" In [3]: df [ "five" ] = df [ "i" ] > 0 In [4]: df Out[4]: ane two iii 4 v a 0.469112 -0.282863 -i.509059 bar True c -1.135632 ane.212112 -0.173215 bar Imitation e 0.119209 -1.044236 -0.861849 bar Truthful f -two.104569 -0.494929 one.071804 bar Imitation h 0.721555 -0.706771 -1.039575 bar Truthful In [v]: df2 = df . reindex ([ "a" , "b" , "c" , "d" , "e" , "f" , "thousand" , "h" ]) In [6]: df2 Out[half dozen]: i two iii 4 five a 0.469112 -0.282863 -1.509059 bar Truthful b NaN NaN NaN NaN NaN c -1.135632 1.212112 -0.173215 bar Fake d NaN NaN NaN NaN NaN east 0.119209 -1.044236 -0.861849 bar True f -2.104569 -0.494929 1.071804 bar False g NaN NaN NaN NaN NaN h 0.721555 -0.706771 -1.039575 bar True To make detecting missing values easier (and across unlike array dtypes), pandas provides the isna() and notna() functions, which are also methods on Serial and DataFrame objects:
In [vii]: df2 [ "one" ] Out[7]: a 0.469112 b NaN c -1.135632 d NaN e 0.119209 f -2.104569 g NaN h 0.721555 Name: one, dtype: float64 In [8]: pd . isna ( df2 [ "one" ]) Out[8]: a Simulated b True c False d True e False f False g True h Fake Name: one, dtype: bool In [nine]: df2 [ "four" ] . notna () Out[ix]: a True b False c True d Fake e True f Truthful g False h True Name: four, dtype: bool In [10]: df2 . isna () Out[ten]: i 2 iii four five a Fake False False Imitation False b True Truthful Truthful True True c Imitation Simulated False Fake False d True True True Truthful True e False Imitation False False False f False False False False False g True True Truthful True True h Simulated Faux False False Imitation Alert
One has to be mindful that in Python (and NumPy), the nan's don't compare equal, only None's practise. Notation that pandas/NumPy uses the fact that np.nan != np.nan , and treats None like np.nan .
In [xi]: None == None # noqa: E711 Out[eleven]: Truthful In [12]: np . nan == np . nan Out[12]: False So as compared to above, a scalar equality comparison versus a None/np.nan doesn't provide useful data.
In [13]: df2 [ "one" ] == np . nan Out[xiii]: a False b False c Fake d Fake e Simulated f False g False h False Name: i, dtype: bool Integer dtypes and missing data¶
Considering NaN is a float, a cavalcade of integers with fifty-fifty one missing values is cast to floating-point dtype (see Support for integer NA for more). pandas provides a nullable integer array, which can be used by explicitly requesting the dtype:
In [fourteen]: pd . Series ([ 1 , 2 , np . nan , 4 ], dtype = pd . Int64Dtype ()) Out[14]: 0 i 1 2 2 <NA> iii 4 dtype: Int64 Alternatively, the string alias dtype='Int64' (notation the capital "I" ) can be used.
See Nullable integer information type for more than.
Datetimes¶
For datetime64[ns] types, NaT represents missing values. This is a pseudo-native sentinel value that can be represented by NumPy in a singular dtype (datetime64[ns]). pandas objects provide compatibility between NaT and NaN .
In [xv]: df2 = df . copy () In [16]: df2 [ "timestamp" ] = pd . Timestamp ( "20120101" ) In [17]: df2 Out[17]: 1 two iii four five timestamp a 0.469112 -0.282863 -1.509059 bar True 2012-01-01 c -one.135632 1.212112 -0.173215 bar False 2012-01-01 e 0.119209 -1.044236 -0.861849 bar Truthful 2012-01-01 f -2.104569 -0.494929 1.071804 bar Faux 2012-01-01 h 0.721555 -0.706771 -1.039575 bar Truthful 2012-01-01 In [18]: df2 . loc [[ "a" , "c" , "h" ], [ "one" , "timestamp" ]] = np . nan In [19]: df2 Out[19]: one two three four five timestamp a NaN -0.282863 -one.509059 bar True NaT c NaN i.212112 -0.173215 bar False NaT east 0.119209 -1.044236 -0.861849 bar True 2012-01-01 f -2.104569 -0.494929 i.071804 bar Simulated 2012-01-01 h NaN -0.706771 -i.039575 bar Truthful NaT In [20]: df2 . dtypes . value_counts () Out[20]: float64 iii object 1 bool 1 datetime64[ns] i dtype: int64 Inserting missing information¶
You can insert missing values past simply assigning to containers. The actual missing value used will exist chosen based on the dtype.
For example, numeric containers will always use NaN regardless of the missing value type chosen:
In [21]: s = pd . Series ([ i , 2 , 3 ]) In [22]: southward . loc [ 0 ] = None In [23]: s Out[23]: 0 NaN 1 two.0 two 3.0 dtype: float64 As well, datetime containers will always utilize NaT .
For object containers, pandas volition utilise the value given:
In [24]: due south = pd . Serial ([ "a" , "b" , "c" ]) In [25]: due south . loc [ 0 ] = None In [26]: s . loc [ 1 ] = np . nan In [27]: s Out[27]: 0 None 1 NaN 2 c dtype: object Calculations with missing data¶
Missing values propagate naturally through arithmetics operations between pandas objects.
In [28]: a Out[28]: one two a NaN -0.282863 c NaN 1.212112 e 0.119209 -1.044236 f -2.104569 -0.494929 h -2.104569 -0.706771 In [29]: b Out[29]: one ii three a NaN -0.282863 -1.509059 c NaN i.212112 -0.173215 due east 0.119209 -1.044236 -0.861849 f -two.104569 -0.494929 ane.071804 h NaN -0.706771 -1.039575 In [30]: a + b Out[30]: one three ii a NaN NaN -0.565727 c NaN NaN 2.424224 e 0.238417 NaN -2.088472 f -four.209138 NaN -0.989859 h NaN NaN -i.413542 The descriptive statistics and computational methods discussed in the data structure overview (and listed hither and here) are all written to account for missing data. For example:
-
When summing data, NA (missing) values will be treated equally zero.
-
If the information are all NA, the outcome will be 0.
-
Cumulative methods like
cumsum()andcumprod()ignore NA values by default, simply preserve them in the resulting arrays. To override this behaviour and include NA values, employskipna=Faux.
In [31]: df Out[31]: 1 2 iii a NaN -0.282863 -1.509059 c NaN i.212112 -0.173215 e 0.119209 -ane.044236 -0.861849 f -2.104569 -0.494929 i.071804 h NaN -0.706771 -1.039575 In [32]: df [ "one" ] . sum () Out[32]: -1.9853605075978744 In [33]: df . hateful ( i ) Out[33]: a -0.895961 c 0.519449 due east -0.595625 f -0.509232 h -0.873173 dtype: float64 In [34]: df . cumsum () Out[34]: one 2 three a NaN -0.282863 -1.509059 c NaN 0.929249 -1.682273 eastward 0.119209 -0.114987 -2.544122 f -i.985361 -0.609917 -1.472318 h NaN -1.316688 -2.511893 In [35]: df . cumsum ( skipna = Faux ) Out[35]: one two three a NaN -0.282863 -ane.509059 c NaN 0.929249 -i.682273 e NaN -0.114987 -ii.544122 f NaN -0.609917 -i.472318 h NaN -1.316688 -2.511893 Sum/prod of empties/nans¶
Warning
This behavior is now standard as of v0.22.0 and is consistent with the default in numpy ; previously sum/prod of all-NA or empty Serial/DataFrames would return NaN. See v0.22.0 whatsnew for more.
The sum of an empty or all-NA Series or column of a DataFrame is 0.
In [36]: pd . Series ([ np . nan ]) . sum () Out[36]: 0.0 In [37]: pd . Series ([], dtype = "float64" ) . sum () Out[37]: 0.0 The product of an empty or all-NA Series or cavalcade of a DataFrame is ane.
In [38]: pd . Series ([ np . nan ]) . prod () Out[38]: 1.0 In [39]: pd . Serial ([], dtype = "float64" ) . prod () Out[39]: i.0 NA values in GroupBy¶
NA groups in GroupBy are automatically excluded. This behavior is consequent with R, for example:
In [twoscore]: df Out[40]: 1 ii three a NaN -0.282863 -1.509059 c NaN one.212112 -0.173215 e 0.119209 -1.044236 -0.861849 f -two.104569 -0.494929 ane.071804 h NaN -0.706771 -ane.039575 In [41]: df . groupby ( "ane" ) . mean () Out[41]: two three 1 -2.104569 -0.494929 ane.071804 0.119209 -1.044236 -0.861849 See the groupby section here for more information.
Cleaning / filling missing data¶
pandas objects are equipped with various data manipulation methods for dealing with missing data.
Filling missing values: fillna¶
fillna() tin can "fill up in" NA values with non-NA data in a couple of ways, which we illustrate:
Replace NA with a scalar value
In [42]: df2 Out[42]: i 2 three iv five timestamp a NaN -0.282863 -1.509059 bar True NaT c NaN 1.212112 -0.173215 bar Simulated NaT eastward 0.119209 -1.044236 -0.861849 bar True 2012-01-01 f -2.104569 -0.494929 1.071804 bar Fake 2012-01-01 h NaN -0.706771 -one.039575 bar Truthful NaT In [43]: df2 . fillna ( 0 ) Out[43]: i two three iv v timestamp a 0.000000 -0.282863 -i.509059 bar True 0 c 0.000000 one.212112 -0.173215 bar False 0 e 0.119209 -ane.044236 -0.861849 bar True 2012-01-01 00:00:00 f -two.104569 -0.494929 1.071804 bar Simulated 2012-01-01 00:00:00 h 0.000000 -0.706771 -1.039575 bar Truthful 0 In [44]: df2 [ "one" ] . fillna ( "missing" ) Out[44]: a missing c missing e 0.119209 f -2.104569 h missing Proper name: one, dtype: object Make full gaps forward or astern
Using the same filling arguments every bit reindexing, we can propagate not-NA values forrard or backward:
In [45]: df Out[45]: one two three a NaN -0.282863 -1.509059 c NaN ane.212112 -0.173215 e 0.119209 -1.044236 -0.861849 f -2.104569 -0.494929 1.071804 h NaN -0.706771 -1.039575 In [46]: df . fillna ( method = "pad" ) Out[46]: ane 2 three a NaN -0.282863 -one.509059 c NaN ane.212112 -0.173215 e 0.119209 -1.044236 -0.861849 f -2.104569 -0.494929 ane.071804 h -two.104569 -0.706771 -1.039575 Limit the amount of filling
If nosotros only desire consecutive gaps filled up to a certain number of data points, we can apply the limit keyword:
In [47]: df Out[47]: i 2 three a NaN -0.282863 -1.509059 c NaN 1.212112 -0.173215 e NaN NaN NaN f NaN NaN NaN h NaN -0.706771 -1.039575 In [48]: df . fillna ( method = "pad" , limit = i ) Out[48]: i two iii a NaN -0.282863 -1.509059 c NaN ane.212112 -0.173215 e NaN one.212112 -0.173215 f NaN NaN NaN h NaN -0.706771 -1.039575 To remind you, these are the available filling methods:
| Method | Activity |
|---|---|
| pad / ffill | Fill values forward |
| bfill / backfill | Fill values backward |
With time series data, using pad/ffill is extremely common then that the "terminal known value" is available at every fourth dimension point.
ffill() is equivalent to fillna(method='ffill') and bfill() is equivalent to fillna(method='bfill')
Filling with a PandasObject¶
You lot can also fillna using a dict or Series that is alignable. The labels of the dict or alphabetize of the Serial must match the columns of the frame you wish to make full. The use case of this is to fill a DataFrame with the mean of that column.
In [49]: dff = pd . DataFrame ( np . random . randn ( x , three ), columns = list ( "ABC" )) In [50]: dff . iloc [ 3 : 5 , 0 ] = np . nan In [51]: dff . iloc [ 4 : 6 , 1 ] = np . nan In [52]: dff . iloc [ 5 : 8 , 2 ] = np . nan In [53]: dff Out[53]: A B C 0 0.271860 -0.424972 0.567020 1 0.276232 -i.087401 -0.673690 2 0.113648 -i.478427 0.524988 three NaN 0.577046 -ane.715002 4 NaN NaN -i.157892 5 -i.344312 NaN NaN half dozen -0.109050 1.643563 NaN seven 0.357021 -0.674600 NaN eight -0.968914 -1.294524 0.413738 9 0.276662 -0.472035 -0.013960 In [54]: dff . fillna ( dff . mean ()) Out[54]: A B C 0 0.271860 -0.424972 0.567020 one 0.276232 -1.087401 -0.673690 two 0.113648 -1.478427 0.524988 3 -0.140857 0.577046 -1.715002 4 -0.140857 -0.401419 -1.157892 5 -one.344312 -0.401419 -0.293543 half dozen -0.109050 1.643563 -0.293543 7 0.357021 -0.674600 -0.293543 8 -0.968914 -ane.294524 0.413738 9 0.276662 -0.472035 -0.013960 In [55]: dff . fillna ( dff . hateful ()[ "B" : "C" ]) Out[55]: A B C 0 0.271860 -0.424972 0.567020 i 0.276232 -1.087401 -0.673690 2 0.113648 -1.478427 0.524988 3 NaN 0.577046 -i.715002 iv NaN -0.401419 -1.157892 5 -1.344312 -0.401419 -0.293543 half dozen -0.109050 1.643563 -0.293543 7 0.357021 -0.674600 -0.293543 eight -0.968914 -1.294524 0.413738 9 0.276662 -0.472035 -0.013960 Same upshot as above, but is aligning the 'fill' value which is a Serial in this case.
In [56]: dff . where ( pd . notna ( dff ), dff . hateful (), axis = "columns" ) Out[56]: A B C 0 0.271860 -0.424972 0.567020 one 0.276232 -1.087401 -0.673690 two 0.113648 -one.478427 0.524988 3 -0.140857 0.577046 -ane.715002 four -0.140857 -0.401419 -1.157892 5 -1.344312 -0.401419 -0.293543 6 -0.109050 1.643563 -0.293543 7 0.357021 -0.674600 -0.293543 8 -0.968914 -1.294524 0.413738 9 0.276662 -0.472035 -0.013960 Dropping axis labels with missing information: dropna¶
You may wish to only exclude labels from a information prepare which refer to missing data. To do this, use dropna() :
In [57]: df Out[57]: one two 3 a NaN -0.282863 -1.509059 c NaN ane.212112 -0.173215 e NaN 0.000000 0.000000 f NaN 0.000000 0.000000 h NaN -0.706771 -1.039575 In [58]: df . dropna ( centrality = 0 ) Out[58]: Empty DataFrame Columns: [one, two, three] Index: [] In [59]: df . dropna ( axis = 1 ) Out[59]: two three a -0.282863 -ane.509059 c ane.212112 -0.173215 e 0.000000 0.000000 f 0.000000 0.000000 h -0.706771 -one.039575 In [60]: df [ "ane" ] . dropna () Out[60]: Series([], Proper name: one, dtype: float64) An equivalent dropna() is available for Series. DataFrame.dropna has considerably more options than Series.dropna, which can be examined in the API.
Interpolation¶
Both Serial and DataFrame objects have interpolate() that, by default, performs linear interpolation at missing data points.
In [61]: ts Out[61]: 2000-01-31 0.469112 2000-02-29 NaN 2000-03-31 NaN 2000-04-28 NaN 2000-05-31 NaN ... 2007-12-31 -6.950267 2008-01-31 -7.904475 2008-02-29 -6.441779 2008-03-31 -eight.184940 2008-04-thirty -9.011531 Freq: BM, Length: 100, dtype: float64 In [62]: ts . count () Out[62]: 66 In [63]: ts . plot () Out[63]: <AxesSubplot:> 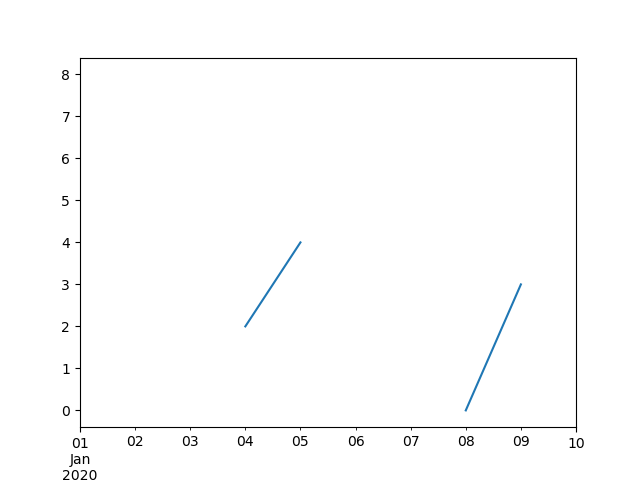
In [64]: ts . interpolate () Out[64]: 2000-01-31 0.469112 2000-02-29 0.434469 2000-03-31 0.399826 2000-04-28 0.365184 2000-05-31 0.330541 ... 2007-12-31 -6.950267 2008-01-31 -7.904475 2008-02-29 -6.441779 2008-03-31 -8.184940 2008-04-30 -9.011531 Freq: BM, Length: 100, dtype: float64 In [65]: ts . interpolate () . count () Out[65]: 100 In [66]: ts . interpolate () . plot () Out[66]: <AxesSubplot:> 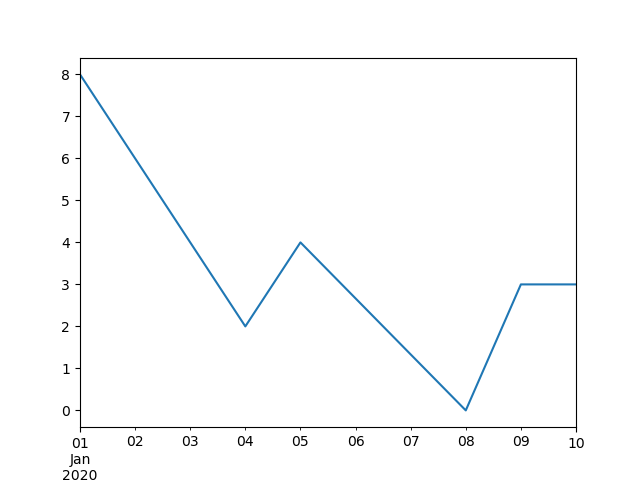
Alphabetize aware interpolation is available via the method keyword:
In [67]: ts2 Out[67]: 2000-01-31 0.469112 2000-02-29 NaN 2002-07-31 -5.785037 2005-01-31 NaN 2008-04-30 -nine.011531 dtype: float64 In [68]: ts2 . interpolate () Out[68]: 2000-01-31 0.469112 2000-02-29 -ii.657962 2002-07-31 -5.785037 2005-01-31 -7.398284 2008-04-30 -ix.011531 dtype: float64 In [69]: ts2 . interpolate ( method = "fourth dimension" ) Out[69]: 2000-01-31 0.469112 2000-02-29 0.270241 2002-07-31 -5.785037 2005-01-31 -vii.190866 2008-04-30 -9.011531 dtype: float64 For a floating-point index, use method='values' :
In [70]: ser Out[70]: 0.0 0.0 1.0 NaN 10.0 x.0 dtype: float64 In [71]: ser . interpolate () Out[71]: 0.0 0.0 1.0 v.0 10.0 x.0 dtype: float64 In [72]: ser . interpolate ( method = "values" ) Out[72]: 0.0 0.0 1.0 1.0 10.0 10.0 dtype: float64 You can also interpolate with a DataFrame:
In [73]: df = pd . DataFrame ( ....: { ....: "A" : [ 1 , ii.1 , np . nan , iv.7 , 5.6 , 6.eight ], ....: "B" : [ 0.25 , np . nan , np . nan , 4 , 12.2 , fourteen.4 ], ....: } ....: ) ....: In [74]: df Out[74]: A B 0 ane.0 0.25 1 ii.1 NaN ii NaN NaN 3 4.seven 4.00 4 5.6 12.20 5 half dozen.8 14.twoscore In [75]: df . interpolate () Out[75]: A B 0 i.0 0.25 i 2.1 1.l 2 3.4 2.75 3 4.vii 4.00 iv v.6 12.20 v 6.viii xiv.40 The method statement gives access to fancier interpolation methods. If you take scipy installed, yous can pass the name of a 1-d interpolation routine to method . Y'all'll desire to consult the total scipy interpolation documentation and reference guide for details. The appropriate interpolation method volition depend on the type of data yous are working with.
-
If you are dealing with a time series that is growing at an increasing rate,
method='quadratic'may be appropriate. -
If yous have values approximating a cumulative distribution function, then
method='pchip'should work well. -
To fill missing values with goal of smoothen plotting, consider
method='akima'.
Warning
These methods require scipy .
In [76]: df . interpolate ( method = "barycentric" ) Out[76]: A B 0 1.00 0.250 1 2.ten -7.660 2 3.53 -4.515 3 four.70 4.000 4 5.60 12.200 5 6.fourscore 14.400 In [77]: df . interpolate ( method = "pchip" ) Out[77]: A B 0 ane.00000 0.250000 1 2.10000 0.672808 two iii.43454 1.928950 3 4.70000 four.000000 4 5.60000 12.200000 5 vi.80000 14.400000 In [78]: df . interpolate ( method = "akima" ) Out[78]: A B 0 one.000000 0.250000 1 2.100000 -0.873316 ii iii.406667 0.320034 3 four.700000 iv.000000 4 5.600000 12.200000 5 6.800000 fourteen.400000 When interpolating via a polynomial or spline approximation, you must also specify the degree or order of the approximation:
In [79]: df . interpolate ( method = "spline" , order = ii ) Out[79]: A B 0 1.000000 0.250000 i two.100000 -0.428598 two 3.404545 1.206900 3 4.700000 4.000000 4 five.600000 12.200000 5 6.800000 14.400000 In [80]: df . interpolate ( method = "polynomial" , order = 2 ) Out[80]: A B 0 1.000000 0.250000 1 2.100000 -2.703846 2 3.451351 -1.453846 three 4.700000 iv.000000 four v.600000 12.200000 5 half dozen.800000 xiv.400000 Compare several methods:
In [81]: np . random . seed ( ii ) In [82]: ser = pd . Series ( np . arange ( one , 10.1 , 0.25 ) ** 2 + np . random . randn ( 37 )) In [83]: missing = np . array ([ four , thirteen , xiv , fifteen , 16 , 17 , eighteen , xx , 29 ]) In [84]: ser [ missing ] = np . nan In [85]: methods = [ "linear" , "quadratic" , "cubic" ] In [86]: df = pd . DataFrame ({ chiliad : ser . interpolate ( method = chiliad ) for m in methods }) In [87]: df . plot () Out[87]: <AxesSubplot:> 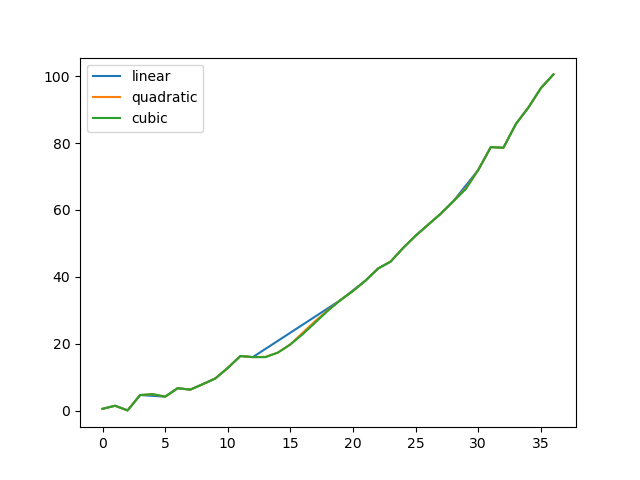
Another utilize case is interpolation at new values. Suppose you have 100 observations from some distribution. And permit's suppose that you lot're specially interested in what's happening around the middle. Y'all can mix pandas' reindex and interpolate methods to interpolate at the new values.
In [88]: ser = pd . Serial ( np . sort ( np . random . uniform ( size = 100 ))) # interpolate at new_index In [89]: new_index = ser . index . union ( pd . Alphabetize ([ 49.25 , 49.5 , 49.75 , fifty.25 , 50.5 , 50.75 ])) In [ninety]: interp_s = ser . reindex ( new_index ) . interpolate ( method = "pchip" ) In [91]: interp_s [ 49 : 51 ] Out[91]: 49.00 0.471410 49.25 0.476841 49.50 0.481780 49.75 0.485998 50.00 0.489266 50.25 0.491814 l.50 0.493995 50.75 0.495763 51.00 0.497074 dtype: float64 Interpolation limits¶
Similar other pandas make full methods, interpolate() accepts a limit keyword argument. Employ this argument to limit the number of sequent NaN values filled since the terminal valid observation:
In [92]: ser = pd . Serial ([ np . nan , np . nan , five , np . nan , np . nan , np . nan , thirteen , np . nan , np . nan ]) In [93]: ser Out[93]: 0 NaN 1 NaN 2 5.0 3 NaN 4 NaN 5 NaN 6 thirteen.0 seven NaN 8 NaN dtype: float64 # make full all consecutive values in a frontward direction In [94]: ser . interpolate () Out[94]: 0 NaN ane NaN 2 five.0 3 seven.0 iv nine.0 5 eleven.0 6 thirteen.0 seven 13.0 8 13.0 dtype: float64 # fill up one sequent value in a frontward direction In [95]: ser . interpolate ( limit = ane ) Out[95]: 0 NaN one NaN 2 5.0 3 vii.0 4 NaN 5 NaN 6 thirteen.0 7 13.0 8 NaN dtype: float64 Past default, NaN values are filled in a forward direction. Use limit_direction parameter to fill backward or from both directions.
# fill one consecutive value backwards In [96]: ser . interpolate ( limit = 1 , limit_direction = "astern" ) Out[96]: 0 NaN 1 five.0 2 5.0 iii NaN 4 NaN five eleven.0 6 13.0 7 NaN 8 NaN dtype: float64 # fill up ane consecutive value in both directions In [97]: ser . interpolate ( limit = one , limit_direction = "both" ) Out[97]: 0 NaN ane 5.0 two five.0 iii seven.0 4 NaN 5 11.0 6 xiii.0 7 13.0 8 NaN dtype: float64 # fill all consecutive values in both directions In [98]: ser . interpolate ( limit_direction = "both" ) Out[98]: 0 v.0 1 5.0 2 v.0 3 7.0 iv nine.0 5 xi.0 vi 13.0 seven thirteen.0 8 thirteen.0 dtype: float64 By default, NaN values are filled whether they are inside (surrounded by) existing valid values, or exterior existing valid values. The limit_area parameter restricts filling to either inside or outside values.
# make full one sequent within value in both directions In [99]: ser . interpolate ( limit_direction = "both" , limit_area = "inside" , limit = 1 ) Out[99]: 0 NaN 1 NaN ii 5.0 3 7.0 4 NaN 5 11.0 6 13.0 seven NaN 8 NaN dtype: float64 # fill all consecutive outside values backward In [100]: ser . interpolate ( limit_direction = "backward" , limit_area = "exterior" ) Out[100]: 0 5.0 1 5.0 2 5.0 3 NaN 4 NaN 5 NaN half-dozen 13.0 seven NaN eight NaN dtype: float64 # fill up all sequent outside values in both directions In [101]: ser . interpolate ( limit_direction = "both" , limit_area = "outside" ) Out[101]: 0 5.0 1 v.0 2 5.0 3 NaN 4 NaN 5 NaN 6 13.0 7 13.0 viii 13.0 dtype: float64 Replacing generic values¶
Often times we want to replace arbitrary values with other values.
supersede() in Serial and supercede() in DataFrame provides an efficient yet flexible way to perform such replacements.
For a Series, you tin can supercede a single value or a list of values by another value:
In [102]: ser = pd . Series ([ 0.0 , 1.0 , two.0 , 3.0 , 4.0 ]) In [103]: ser . supersede ( 0 , five ) Out[103]: 0 5.0 ane one.0 2 2.0 iii 3.0 four 4.0 dtype: float64 Y'all can replace a listing of values by a listing of other values:
In [104]: ser . supervene upon ([ 0 , 1 , 2 , three , 4 ], [ 4 , three , 2 , i , 0 ]) Out[104]: 0 iv.0 ane 3.0 2 ii.0 3 1.0 iv 0.0 dtype: float64 You can likewise specify a mapping dict:
In [105]: ser . replace ({ 0 : 10 , 1 : 100 }) Out[105]: 0 x.0 1 100.0 ii 2.0 3 3.0 4 4.0 dtype: float64 For a DataFrame, you can specify individual values by cavalcade:
In [106]: df = pd . DataFrame ({ "a" : [ 0 , ane , 2 , 3 , 4 ], "b" : [ 5 , 6 , seven , 8 , 9 ]}) In [107]: df . replace ({ "a" : 0 , "b" : v }, 100 ) Out[107]: a b 0 100 100 one ane 6 2 2 seven 3 three viii 4 iv 9 Instead of replacing with specified values, you can care for all given values equally missing and interpolate over them:
In [108]: ser . replace ([ 1 , 2 , 3 ], method = "pad" ) Out[108]: 0 0.0 1 0.0 2 0.0 three 0.0 4 iv.0 dtype: float64 String/regular expression replacement¶
Notation
Python strings prefixed with the r character such every bit r'hullo world' are and then-called "raw" strings. They have different semantics regarding backslashes than strings without this prefix. Backslashes in raw strings volition be interpreted every bit an escaped backslash, e.chiliad., r'\' == '\\' . Y'all should read near them if this is unclear.
Replace the '.' with NaN (str -> str):
In [109]: d = { "a" : list ( range ( iv )), "b" : list ( "ab.." ), "c" : [ "a" , "b" , np . nan , "d" ]} In [110]: df = pd . DataFrame ( d ) In [111]: df . replace ( "." , np . nan ) Out[111]: a b c 0 0 a a 1 one b b 2 ii NaN NaN iii iii NaN d At present do it with a regular expression that removes surrounding whitespace (regex -> regex):
In [112]: df . replace ( r "\s*\.\due south*" , np . nan , regex = True ) Out[112]: a b c 0 0 a a 1 1 b b ii two NaN NaN 3 3 NaN d Replace a few dissimilar values (list -> listing):
In [113]: df . replace ([ "a" , "." ], [ "b" , np . nan ]) Out[113]: a b c 0 0 b b 1 1 b b ii 2 NaN NaN 3 three NaN d list of regex -> list of regex:
In [114]: df . supercede ([ r "\." , r "(a)" ], [ "dot" , r "\1stuff" ], regex = True ) Out[114]: a b c 0 0 astuff astuff i 1 b b 2 2 dot NaN 3 iii dot d Only search in column 'b' (dict -> dict):
In [115]: df . supplant ({ "b" : "." }, { "b" : np . nan }) Out[115]: a b c 0 0 a a one 1 b b 2 2 NaN NaN three three NaN d Same every bit the previous instance, but use a regular expression for searching instead (dict of regex -> dict):
In [116]: df . replace ({ "b" : r "\s*\.\s*" }, { "b" : np . nan }, regex = True ) Out[116]: a b c 0 0 a a 1 ane b b 2 ii NaN NaN 3 3 NaN d You can pass nested dictionaries of regular expressions that use regex=Truthful :
In [117]: df . supervene upon ({ "b" : { "b" : r "" }}, regex = True ) Out[117]: a b c 0 0 a a 1 one b 2 2 . NaN 3 3 . d Alternatively, you can pass the nested dictionary like and so:
In [118]: df . replace ( regex = { "b" : { r "\south*\.\s*" : np . nan }}) Out[118]: a b c 0 0 a a 1 one b b 2 2 NaN NaN iii 3 NaN d You lot tin can also use the group of a regular expression match when replacing (dict of regex -> dict of regex), this works for lists as well.
In [119]: df . replace ({ "b" : r "\s*(\.)\due south*" }, { "b" : r "\1ty" }, regex = True ) Out[119]: a b c 0 0 a a one 1 b b 2 2 .ty NaN iii 3 .ty d Yous can pass a list of regular expressions, of which those that match will be replaced with a scalar (list of regex -> regex).
In [120]: df . supercede ([ r "\s*\.\s*" , r "a|b" ], np . nan , regex = Truthful ) Out[120]: a b c 0 0 NaN NaN i 1 NaN NaN 2 two NaN NaN three 3 NaN d All of the regular expression examples can also exist passed with the to_replace argument equally the regex argument. In this case the value argument must be passed explicitly by proper name or regex must be a nested dictionary. The previous example, in this example, would and then be:
In [121]: df . replace ( regex = [ r "\s*\.\s*" , r "a|b" ], value = np . nan ) Out[121]: a b c 0 0 NaN NaN 1 1 NaN NaN two ii NaN NaN 3 3 NaN d This can be convenient if you do not want to pass regex=True every time you lot want to apply a regular expression.
Annotation
Anywhere in the above supplant examples that you run into a regular expression a compiled regular expression is valid also.
Numeric replacement¶
supervene upon() is similar to fillna() .
In [122]: df = pd . DataFrame ( np . random . randn ( 10 , 2 )) In [123]: df [ np . random . rand ( df . shape [ 0 ]) > 0.v ] = 1.five In [124]: df . replace ( ane.5 , np . nan ) Out[124]: 0 1 0 -0.844214 -one.021415 one 0.432396 -0.323580 two 0.423825 0.799180 iii 1.262614 0.751965 four NaN NaN 5 NaN NaN half dozen -0.498174 -1.060799 7 0.591667 -0.183257 8 i.019855 -1.482465 nine NaN NaN Replacing more than one value is possible past passing a list.
In [125]: df00 = df . iloc [ 0 , 0 ] In [126]: df . replace ([ 1.5 , df00 ], [ np . nan , "a" ]) Out[126]: 0 ane 0 a -1.021415 1 0.432396 -0.323580 2 0.423825 0.799180 iii 1.262614 0.751965 4 NaN NaN v NaN NaN vi -0.498174 -1.060799 7 0.591667 -0.183257 8 1.019855 -ane.482465 9 NaN NaN In [127]: df [ 1 ] . dtype Out[127]: dtype('float64') You tin also operate on the DataFrame in place:
In [128]: df . replace ( i.5 , np . nan , inplace = Truthful ) Missing information casting rules and indexing¶
While pandas supports storing arrays of integer and boolean type, these types are not capable of storing missing data. Until nosotros can switch to using a native NA type in NumPy, nosotros've established some "casting rules". When a reindexing operation introduces missing information, the Series will exist bandage according to the rules introduced in the table below.
| data type | Bandage to |
|---|---|
| integer | bladder |
| boolean | object |
| bladder | no cast |
| object | no bandage |
For case:
In [129]: s = pd . Series ( np . random . randn ( 5 ), index = [ 0 , ii , 4 , half-dozen , seven ]) In [130]: southward > 0 Out[130]: 0 True 2 True four Truthful half-dozen Truthful 7 Truthful dtype: bool In [131]: ( s > 0 ) . dtype Out[131]: dtype('bool') In [132]: crit = ( south > 0 ) . reindex ( listing ( range ( 8 ))) In [133]: crit Out[133]: 0 True 1 NaN ii True 3 NaN four True five NaN 6 True 7 True dtype: object In [134]: crit . dtype Out[134]: dtype('O') Unremarkably NumPy will complain if yous effort to use an object array (fifty-fifty if it contains boolean values) instead of a boolean array to get or set values from an ndarray (e.1000. selecting values based on some criteria). If a boolean vector contains NAs, an exception will be generated:
In [135]: reindexed = south . reindex ( list ( range ( eight ))) . fillna ( 0 ) In [136]: reindexed [ crit ] --------------------------------------------------------------------------- ValueError Traceback (near recent phone call terminal) Input In [ 136 ], in < module > ----> i reindexed [ crit ] File /pandas/pandas/core/serial.py:979, in Serial.__getitem__ (cocky, key) 976 if is_iterator ( key ): 977 key = list ( key ) --> 979 if com . is_bool_indexer ( primal ): 980 fundamental = check_bool_indexer ( self . alphabetize , key ) 981 key = np . asarray ( fundamental , dtype = bool ) File /pandas/pandas/cadre/mutual.py:144, in is_bool_indexer (key) 140 na_msg = "Cannot mask with not-boolean array containing NA / NaN values" 141 if lib . infer_dtype ( central ) == "boolean" and isna ( key ) . whatever (): 142 # Don't heighten on eastward.g. ["A", "B", np.nan], see 143 # test_loc_getitem_list_of_labels_categoricalindex_with_na --> 144 raise ValueError ( na_msg ) 145 return False 146 return Truthful ValueError: Cannot mask with non-boolean array containing NA / NaN values Withal, these can be filled in using fillna() and it will work fine:
In [137]: reindexed [ crit . fillna ( False )] Out[137]: 0 0.126504 ii 0.696198 four 0.697416 6 0.601516 vii 0.003659 dtype: float64 In [138]: reindexed [ crit . fillna ( True )] Out[138]: 0 0.126504 1 0.000000 2 0.696198 3 0.000000 4 0.697416 5 0.000000 6 0.601516 7 0.003659 dtype: float64 pandas provides a nullable integer dtype, but you lot must explicitly request information technology when creating the series or column. Notice that nosotros use a capital "I" in the dtype="Int64" .
In [139]: s = pd . Series ([ 0 , 1 , np . nan , 3 , four ], dtype = "Int64" ) In [140]: s Out[140]: 0 0 ane 1 two <NA> three 3 4 4 dtype: Int64 See Nullable integer data blazon for more.
Experimental NA scalar to denote missing values¶
Alarm
Experimental: the behaviour of pd.NA can still change without alarm.
New in version ane.0.0.
Starting from pandas 1.0, an experimental pd.NA value (singleton) is available to correspond scalar missing values. At this moment, it is used in the nullable integer, boolean and defended string data types every bit the missing value indicator.
The goal of pd.NA is provide a "missing" indicator that can exist used consistently beyond data types (instead of np.nan , None or pd.NaT depending on the information type).
For example, when having missing values in a Serial with the nullable integer dtype, it volition use pd.NA :
In [141]: s = pd . Series ([ i , 2 , None ], dtype = "Int64" ) In [142]: southward Out[142]: 0 one 1 2 two <NA> dtype: Int64 In [143]: south [ 2 ] Out[143]: <NA> In [144]: s [ 2 ] is pd . NA Out[144]: Truthful Currently, pandas does not yet utilize those data types by default (when creating a DataFrame or Serial, or when reading in data), so you need to specify the dtype explicitly. An easy style to convert to those dtypes is explained here.
Propagation in arithmetic and comparison operations¶
In general, missing values propagate in operations involving pd.NA . When one of the operands is unknown, the issue of the operation is also unknown.
For example, pd.NA propagates in arithmetic operations, similarly to np.nan :
In [145]: pd . NA + 1 Out[145]: <NA> In [146]: "a" * pd . NA Out[146]: <NA> There are a few special cases when the event is known, even when i of the operands is NA .
In [147]: pd . NA ** 0 Out[147]: 1 In [148]: one ** pd . NA Out[148]: i In equality and comparison operations, pd.NA also propagates. This deviates from the behaviour of np.nan , where comparisons with np.nan always render Imitation .
In [149]: pd . NA == 1 Out[149]: <NA> In [150]: pd . NA == pd . NA Out[150]: <NA> In [151]: pd . NA < ii.5 Out[151]: <NA> To bank check if a value is equal to pd.NA , the isna() role can be used:
In [152]: pd . isna ( pd . NA ) Out[152]: True An exception on this basic propagation rule are reductions (such as the mean or the minimum), where pandas defaults to skipping missing values. See above for more.
Logical operations¶
For logical operations, pd.NA follows the rules of the 3-valued logic (or Kleene logic, similarly to R, SQL and Julia). This logic means to only propagate missing values when it is logically required.
For example, for the logical "or" operation ( | ), if i of the operands is Truthful , we already know the event will exist True , regardless of the other value (then regardless the missing value would be True or False ). In this case, pd.NA does not propagate:
In [153]: True | Imitation Out[153]: True In [154]: True | pd . NA Out[154]: True In [155]: pd . NA | Truthful Out[155]: Truthful On the other mitt, if one of the operands is Simulated , the result depends on the value of the other operand. Therefore, in this case pd.NA propagates:
In [156]: False | True Out[156]: Truthful In [157]: False | False Out[157]: Fake In [158]: Simulated | pd . NA Out[158]: <NA> The behaviour of the logical "and" operation ( & ) can exist derived using similar logic (where now pd.NA will not propagate if 1 of the operands is already Fake ):
In [159]: False & True Out[159]: False In [160]: False & False Out[160]: Fake In [161]: Faux & pd . NA Out[161]: Imitation In [162]: True & True Out[162]: True In [163]: True & False Out[163]: False In [164]: Truthful & pd . NA Out[164]: <NA> NA in a boolean context¶
Since the bodily value of an NA is unknown, it is cryptic to convert NA to a boolean value. The following raises an fault:
In [165]: bool ( pd . NA ) --------------------------------------------------------------------------- TypeError Traceback (most recent call concluding) Input In [ 165 ], in < module > ----> 1 bool ( pd . NA ) File /pandas/pandas/_libs/missing.pyx:382, in pandas._libs.missing.NAType.__bool__ () TypeError: boolean value of NA is cryptic This also means that pd.NA cannot be used in a context where it is evaluated to a boolean, such as if condition: ... where condition can potentially be pd.NA . In such cases, isna() can exist used to check for pd.NA or condition existence pd.NA can be avoided, for example by filling missing values beforehand.
A similar situation occurs when using Series or DataFrame objects in if statements, meet Using if/truth statements with pandas.
NumPy ufuncs¶
pandas.NA implements NumPy'southward __array_ufunc__ protocol. Well-nigh ufuncs work with NA , and generally return NA :
In [166]: np . log ( pd . NA ) Out[166]: <NA> In [167]: np . add together ( pd . NA , 1 ) Out[167]: <NA> Warning
Currently, ufuncs involving an ndarray and NA will render an object-dtype filled with NA values.
In [168]: a = np . array ([ ane , 2 , three ]) In [169]: np . greater ( a , pd . NA ) Out[169]: array([<NA>, <NA>, <NA>], dtype=object) The return type here may modify to render a different array type in the time to come.
See DataFrame interoperability with NumPy functions for more on ufuncs.
Conversion¶
If you have a DataFrame or Serial using traditional types that accept missing data represented using np.nan , at that place are convenience methods convert_dtypes() in Series and convert_dtypes() in DataFrame that tin can catechumen data to use the newer dtypes for integers, strings and booleans listed here. This is especially helpful after reading in data sets when letting the readers such equally read_csv() and read_excel() infer default dtypes.
In this example, while the dtypes of all columns are changed, we evidence the results for the beginning x columns.
In [170]: bb = pd . read_csv ( "data/baseball game.csv" , index_col = "id" ) In [171]: bb [ bb . columns [: x ]] . dtypes Out[171]: player object twelvemonth int64 stint int64 team object lg object thou int64 ab int64 r int64 h int64 X2b int64 dtype: object In [172]: bbn = bb . convert_dtypes () In [173]: bbn [ bbn . columns [: ten ]] . dtypes Out[173]: player string year Int64 stint Int64 team string lg string g Int64 ab Int64 r Int64 h Int64 X2b Int64 dtype: object Source: https://pandas.pydata.org/pandas-docs/stable/user_guide/missing_data.html
0 Response to "Nicolaescu, L.: the Index of Families of Boundary Value Problems for Dirac Operators"
Post a Comment